Introduction
Ensuring the reliability of published research has become increasingly important to publishers, funders, institutions, and others over the past decade. This can mostly be attributed to increased recognition of the many factors that affect the quality and credibility of the research and publishing process. Some of the main challenges to publishing reproducible research are steeped in the research process itself, such as underpowered study design, while others are due to inadequate descriptions of methods and materials, the selective presentation of results, or even the deep-rooted practice and norms in assessing published research, which could result in publication bias.1
When surveyed, authors of Nature-branded journals identify three key constituents with the greatest potential to improve the reproducibility of published research: researchers, laboratory heads, and publishers.2 So, what can publishers and editors do to ensure that research published in their journals can be reproduced by others? Here, we discuss three approaches we have taken at our journals. These approaches exemplify a range of ways in which publishers can add value to the peer review process and to the published article, and provide an essential publishing infrastructure to support reproducible and open research practice. Each approach seeks to address a specific constellation of issues, and may be better suited to some kinds of research than others. The approaches we discuss are:
- Introducing a checklist for transparent reporting in life science articles,
- Supporting computational reproducibility through peer review of code, and
- Registered reports, an innovative article format aiming to reduce publication bias.
Below we outline our experience with these initiatives and how the research community has responded.
Checklists for Transparent Reporting in Life Sciences
In 2013, the Nature-branded journals announced a set of measures3 intended to support publication of reproducible research. A central feature of this effort was to introduce a mandatory reporting checklist for all primary research life science papers published in Nature-branded journals. The reporting checklist summarized important aspects of experimental design, methodology, and analysis that are considered to underlie irreproducibility and increase bias in reporting research findings, particularly of preclinical animal research.4 It also includes information on statistics, materials, data and code availability, and in-lab replicability. The checklist is made available to reviewers during the peer review process and author compliance is monitored by journal editors.
Inspired by the pioneering work of the EQUATOR5 network in raising transparency and reporting standards6 in clinical research, we hoped the checklist would raise the standard of reporting in published life science research articles in our journals. A second, more aspirational long-term goal was that the checklist may spur changes in researcher and laboratory practice.
Impact of the checklist on reporting and researcher perceptions
Independent studies7,8 show that the checklist has had an impact on transparency of reporting in both published articles and on laboratory practice. Assessment of life science articles from Nature-branded journals found a marked improvement in the reporting of randomization, blinding, exclusions, sample size calculation for in vivo research, and statistics with a far more modest impact on incorporation of these elements into experimental study design.7
Authors have also reported an impact of the checklist on statistics reporting: 83% of respondents in a survey of our authors felt that using the checklist had significantly improved the reporting of statistics within the published papers. The checklist was also found to help increase data deposition and improve description of reagents (Figure 1).2
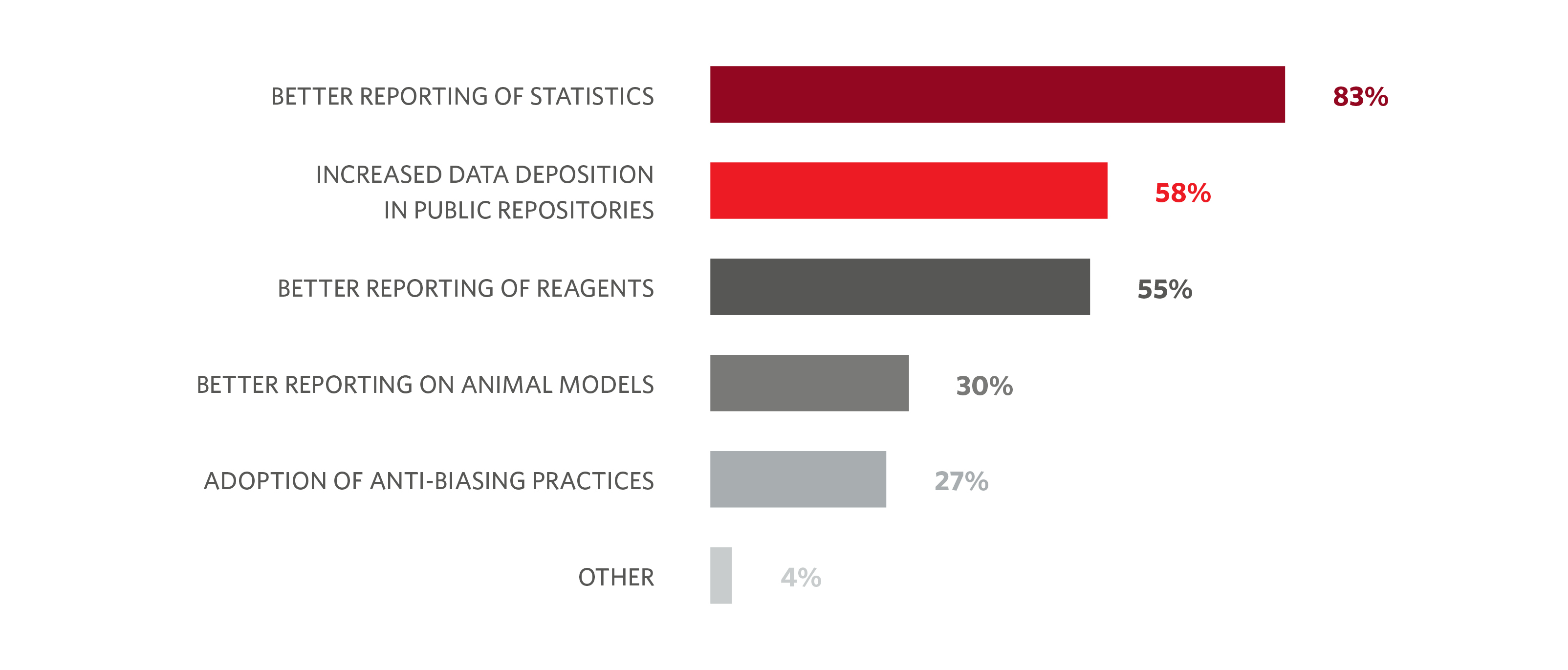
Many first-time submitters to Nature-branded journals only consider using the checklist after submission of the first draft of their manuscript. But the checklist nevertheless appears to have made an impact on laboratory practice. Approximately a quarter of researchers we surveyed report using the checklist to a large extent beyond the journal publishing process, while 78% of respondents said they use the checklist in this way to a small extent.2
Completing a checklist undoubtedly costs researchers more time and effort. But in our experience, author feedback has been largely positive and authors acknowledge the benefits of transparency and a structured set of requirements in improving the manuscript.
Not all feedback from authors has been positive. Some tell us that while the checklist is right in intent, it is too generic to be useful across the broad swath of life science papers. We have begun making in-roads to complementing the basic checklist with the development of more detailed methods-specific requirements9 and broader policies on data, code, materials, and protocol availability.
Lessons learned and next steps
The life science reporting checklist has now become an essential operational tool, allowing us to present editorial policy requirements in a consolidated, accessible manner and easing the challenges of policy compliance for authors, reviewers, editors and others.
The success of the checklist approach in the life sciences was contingent on making the checklist mandatory together with a strong editorial commitment to monitoring compliance. Although assessing compliance can be a resource-intensive, and sometimes frustrating, process for authors and editors, it was absolutely necessary to realizing the benefits of the checklist.
The success of the checklist approach in the life sciences was contingent on making the checklist mandatory together with a strong editorial commitment to monitoring compliance.
As a next step in the development and implementation of reporting checklists, we are working with a cross-publisher group of journal editors and experts in transparency and reproducibility to define a “minimum standards” framework and checklist for reporting across four main areas: Materials, Design, Analysis, and Reporting (MDAR).10 We believe publishers and other stakeholders agreeing on a minimum set of reporting standards and recommendations will help simplify the diverse range of policies and expectations for researchers. Broad uniformity will reinforce standards of reporting, raise awareness early in the life cycle of a study, and help move the field toward greater rigor and transparency in reporting.
Supporting Computational Reproducibility Through Peer Review of Code
Beyond the findings they report, scientific papers are sources of data, code, methodological information, and protocols. In fact, this material forms the building blocks for all future scientific projects and discoveries that a paper may inspire and are essential for reproducibility of the findings. Authors expect their article to be reviewed by peers; why should these other key elements also not meet the same quality assessments?
More than a decade ago, Nature Methods started to require authors of papers in which new code was central to the main paper, to submit the code (preferably as source code) so that the code could be checked by the reviewers. During code peer review, reviewers were asked to verify that the code was functional and “ran as advertised” and that the author’s analyses using the code were correct. Authors were also required to share code so that it could be readily used by the academic community. In those early days, the code was provided in the final paper in a folder that was part of the supplementary information or more recently, as a link to a GitHub11 folder or similar.12
Implementing code peer review
Over the years, several other Nature-branded journals have adopted the practice of peer reviewing code and this experience has made it clear that peer reviewing code is cumbersome for all parties involved. In order to peer review code, authors have to compile it in a format that is accessible to others. Reviewers need to be able to download the code and data, and then set up their own suitable computational environment, often requiring them to install the many dependencies needed to make it all work, and use their own servers to run and validate the code. Even using services like GitHub,11 Zenodo,13 or Figshare14 to check whether code works as advertised and whether it is properly documented and accessible is time-intensive and can challenge the anonymity of reviewers (Figure 2).
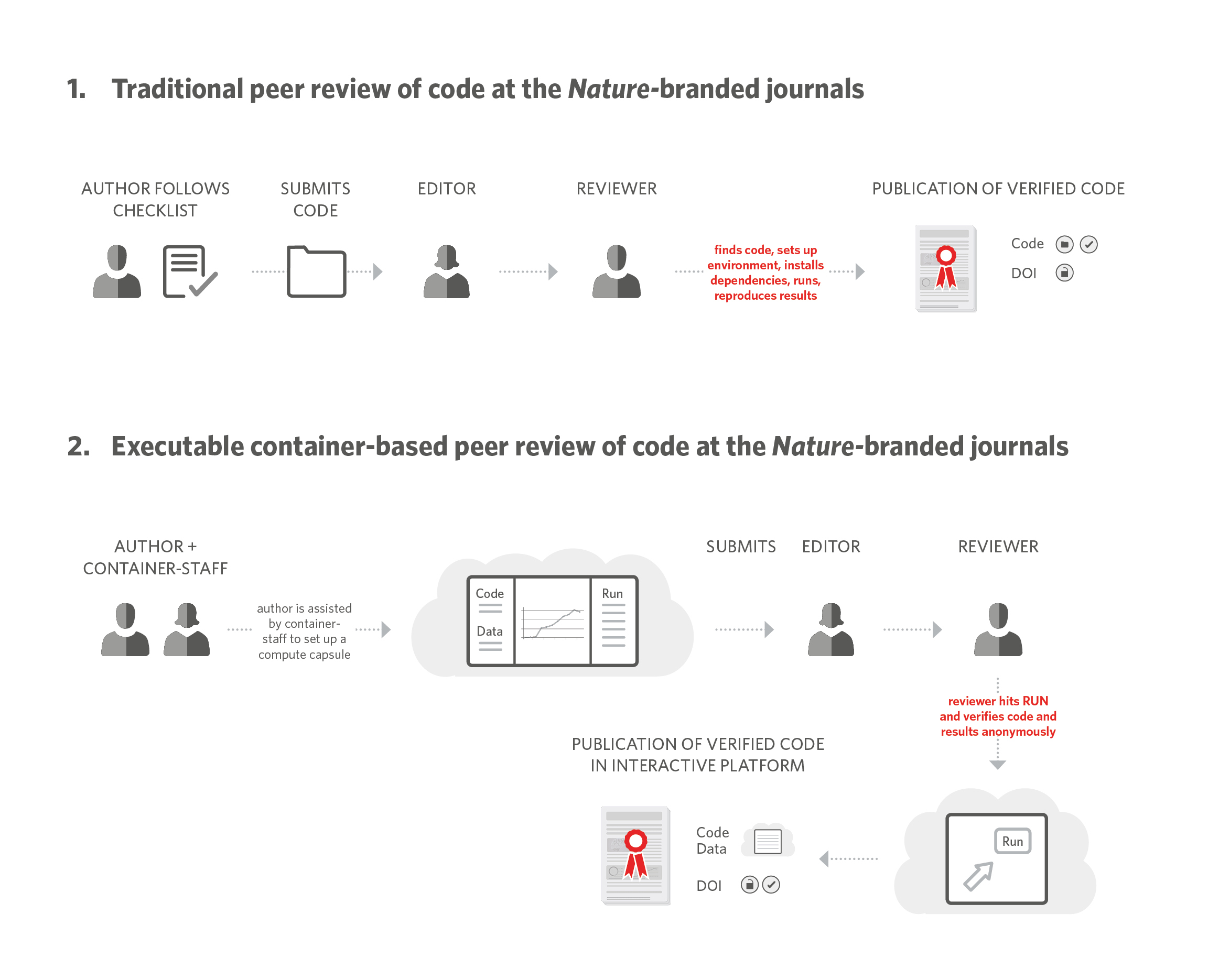
In 2018, we formalized guidelines15 to help authors, editors, and reviewers during code peer review. As part of the submission process, authors are asked to fill a “Software and Code submission checklist” that is used by the editors and reviewers to ensure all the necessary information and documentation to help a third party find, install, and run the code is provided. Ultimately, these documents have facilitated code peer review and the practice is being adopted at more of our journals as a result.16
Container-based peer review
Over the last few years, a number of container-based platforms capable of bundling code, data, and computing environments into a single platform have entered the market. These interactive capsules, or notebooks, make it easier to navigate source code and data, and reproduce the results by enabling the rerun of analyses with a simple click of a button. These products allow code to be included in the published paper as an interactive, reproducible capsule.
We became interested in using these container solutions for submission and peer review of code. In August 2018, we launched a trial to test the use of Code Ocean’s container based platform for peer review and publication of code at several Nature-branded journals.17
Working with Code Ocean, we developed workflows and functionality that enable authors to submit their code and data and compile it into a “compute capsule” which is then used by the editors during peer review. The compute capsule is accessed anonymously by the reviewers who can then run the code to reproduce the analysis and results without needing to install any software. Reviewers are provided ample time to run the code in the cloud for its verification. Upon publication, the capsule is given a DOI and provided as an open platform to all readers for verification, and use (Figure 2).
Lessons learned and next steps
Although container-based peer review is not a solution for every paper with custom code, particularly those that require very large datasets or extremely long running times, and there can be barriers to sharing complex code, feedback from our trial indicates containers improve the quality, documentation, and accessibility of software for both reviewers and users. These new tools also facilitate compliance with the journal’s policies and practices, and ensure higher reproducibility of the research presented in the article. This also benefits reviewers and authors by improving the peer review experience and supporting the sharing of code that is reproducible as well as useful.
We are deeply invested in improving the quality of research reported in our papers, and that includes the elements associated with it. Encouraging code submission, peer review, and publication in open, interactive platforms is one of several important steps we will continue to take to ensure published research is more than a report of the findings.
Registered Reports, an Innovative Article Format to Promote Methodological Rigor and Reduce Publication Bias
Positive or statistically significant findings are much more likely to be published than null or negative findings.18 Such publication bias undermines the credibility of science and its ability to self-correct. In addition to publication bias, we know from studies in meta-science that the traditional peer review and publication model enables questionable research practices (e.g., p-hacking and hypothesizing after the results are known), which compromise the validity and trustworthiness of science. Further, the scientific record and process also suffers when journals and authors place outsized focus on novel results rather than methodological rigor.
Publication bias, questionable research practices, and an outsized emphasis on novel results have all contributed to substantial waste in research—which, according to one estimate, would be as high as 85% in the biomedical sciences.19
One approach to reducing waste in research while tackling questionable research practices and neutralizing publication bias is through an innovative article format called Registered Reports. With Registered Reports, decisions for acceptance are made before the data are collected or analyzed, shifting the emphasis from the results of research (which are beyond scientists’ control) to the importance of the research question and the rigor of the methodology.
Registered Reports in their current form were introduced at the journal Cortex in 2013, although a precursor format was used in The Lancet from 1997 to 2015. Registered Reports are currently offered by approximately 200 journals20 including Nature Human Behaviour, a Nature-branded journal. Nature Human Behaviour adopted the format at the journal’s launch in 2017 and was the first highly-selective journal to offer the format.20
How Registered Reports work
The key distinguishing feature of the Registered Report format is its two-stage peer review system (Figure 3).
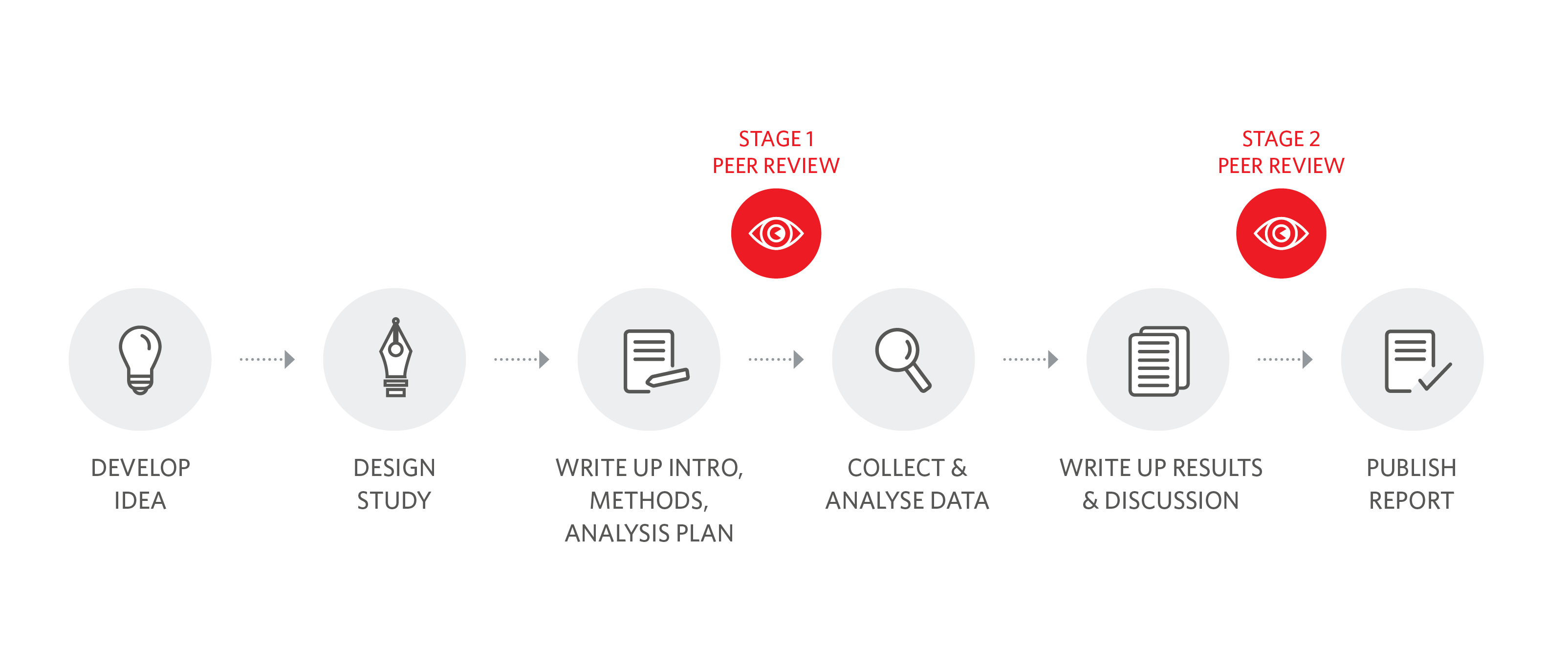
In the first stage, researchers put together their research protocol and write up their introduction, methods, and analysis plan (including any pilot data). This Stage 1 Registered Report is submitted for peer review and evaluated on the basis of the importance of the research question and the rigor of the methodology. If editors and reviewers are satisfied that the protocol meets the journal’s criteria and is methodologically highly robust, the Stage 1 submission is accepted in principle for publication. The authors then collect their data, analyze them, and write up their Stage 2 Registered Report submission, which includes the accepted protocol plus the results and discussion. The full paper is peer reviewed again, but reviewers are only asked to comment on whether the authors adhered to their accepted protocol and if their conclusions are supported by the data. The novelty, conclusiveness, or direction of results is irrelevant for making a decision on publication. If the authors followed their protocol and their conclusions are sensible, the Stage 2 submission is accepted for publication.
Traditional peer review takes place after the research has been conducted—frequently in the form of a postmortem (what did the “patient” die of?). By contrast, the Registered Report peer review model is designed to ensure that research projects are as strong as they can possibly be before substantial resources are invested in data collection, hence preventing unnecessary waste. Because commitment to publish is made at a time when the results are not known, Registered Reports help neutralize publication bias—journals commit to publishing a piece of research regardless of the direction of the results. Finally, because at the time of Stage 2 submission authors are held to their accepted protocol and are required to clearly distinguish between registered analyses and exploratory analyses, questionable research practices are minimized. In fact, incentives for them have been essentially removed because research will be published regardless of the results.
Impact and lessons learned
Registered Reports are not a panacea nor are they suitable for all types of research. Science advances through both discovery and (dis)confirmation. Discovery science involves exploring the full space of possibilities, learning from trial-and-error, and would therefore be crippled by limits to exploration. This means that Registered Reports are not well-suited for exploratory, discovery science. On the other hand, hypothesis-driven research, which aims to (dis)confirm existing theories and predictions, proceeds from a pre-existing set of priors to determine whether hypotheses and predictions are confirmed by the data. For confirmatory research to be valid, it needs to be based on a prespecified and fixed set of hypotheses that is immune to arbitrary researcher degrees of freedom in analyses. Registered Reports are ideally suited for confirmatory research.
Since 2017, Nature Human Behaviour has accepted 11 Stage 1 Registered Reports and has published two Stage 2 submissions. All Stage 1 Registered Reports accepted by the journal and currently made publicly available by their authors can be found on figshare.21 The published Stage 2 submissions, along with other related content on Registered Reports, can be found at https://www.nature.com/collections/cjjiifhaff.22 The journal is committed to promoting the format and encouraging scientists to adopt it for their hypothesis-driven research. Our philosophy is that, if the question is important and the methods are robust and rigorous, the answer will be important, no matter what it is.
Registered Reports represent a radically different way of doing and publishing confirmatory research—and the adoption of the format isn’t without challenges. Authors need to invest more time in the development of their project upfront and to acquire stronger experimental design and statistical expertise (for instance, the fundamentals of a priori sample size specification). Editors and reviewers need to use different criteria than those used to evaluate “standard” submissions.23 And all involved need to develop an entirely different approach to what matters in science. Although the learning curve for everybody involved is steep, it represents a worthwhile investment that has the potential to substantially increase scientific credibility.
Conclusions
While journal editors and publishers must play their part in promoting transparency and reproducibility, meaningful, sustained impact that reinforces open research practices through mentoring, training, and the research process can only come from multiple stakeholders. Institutions and funders, in particular, will need to provide much needed support in training, mentoring, and infrastructure including resource and support for managing the underlying outputs of the research, data, code, materials, and protocols
Acknowledgements
The 2017 reproducibility survey with Nature journal authors was carried out by Gregory Goodey, the graphics were produced by Meredith Simonds, and Elizabeth Hawkins provided support with developmental editing.
Author contributions
The author list is ordered alphabetically; all authors contributed to the original draft preparation, review, and editing. Sowmya Swaminathan is the corresponding author.
Competing interests
The authors declare no competing interest and no funding source. All authors are employees of Springer Nature.
References and Links
- Munafò MR, Nosek BA, Bishop DVM, et al. A manifesto for reproducible science. Nature Hum Behav 2017;1:2–9. https://doi.org/10.1038/s41562-016-0021.
- Nature Reproducibility Survey 2017. figshare. https://doi.org/10.6084/m9.figshare.6139937.v4.
- Announcement: Reducing our irreproducibility. Nature 2013;496:398. https://doi.org/10.1038/496398a.
- Landis SC, Amara SG, Asadullah K, et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature 2012;490:187–191. https://doi.org/10.1038/nature11556.
- http://www.equator-network.org/about-us/equator-network-what-we-do-and-how-we-are-organised/
- Simera I, Altman DG, Moher D, Schulz KF, Hoey J. Guidelines for reporting health research: The EQUATOR Network’s Survey of Guideline Authors. PLoS Med 2008;5:e139. https://doi.org/10.1371/journal.pmed.0050139.
- The NPQIP Collaborative Group. Did a change in Nature journals’ editorial policy for life sciences research improve reporting? BMJ Open Sci 2019;3:e000035. https://doi.org/10.1136/bmjos-2017-000035.
- Han S, Olonisakin TF, Pribis JP, et al. A checklist is associated with increased quality of reporting preclinical biomedical research: A systematic review. PLoS ONE 2007;12(9):e0183591. https://doi.org/10.1371/journal.pone.0183591.
- Announcement: Towards greater reproducibility for life sciences research in Nature 2017;546:8 https://doi.org/10.1038/546008a.
- Chambers K, Collings A, Graf C, et al. Towards minimum reporting standards for life scientists. Available at: https://osf.io/preprints/metaarxiv/9sm4x.
- https://github.com/
- Social software. Nature Methods2007;4: https://doi.org/10.1038/nmeth0307-189.
- https://zenodo.org/
- https://figshare.com/
- https://www.nature.com/documents/GuidelinesCodePublication.pdf
- Does your code stand up to scrutiny? Nature2018;555: https://doi.org/10.1038/d41586-018-02741-4.
- Nature Research journals trial new tools to enhance code peer review and publication. Available at: http://blogs.nature.com/ofschemesandmemes/2018/08/01/nature-research-journals-trial-new-tools-to-enhance-code-peer-review-and-publication.
- Fanelli D. Negative results are disappearing from most disciplines and countries. Scientometrics 2012;90:891–904. https://doi.org/10.1007/s11192-011-0494-7.
- Chalmers I, Glasziou P. Avoidable waste in the production and reporting of research evidence. Lancet. 2009;374(9683):86–89. https://doi.org/10.1016/S0140-6736(09)60329-9.
- https://cos.io/rr/
- Discover research from Registered Reports at Nature Human Behaviour. Available at: https://springernature.figshare.com/registered-reports_NHB.
- https://www.nature.com/collections/cjjiifhaff
- https://socialsciences.nature.com/users/174171-marike-schiffer/posts/45906-publishing-registered-reports
Stavroula Kousta (@KoustaStavroula) is Chief Editor, Nature Human Behaviour, Erika Pastrana (@ErikaPastrana1) is Editorial Director, Applied & Chemical Sciences Nature Journals), and Sowmya Swaminathan (@SowmyaSwaminat1) is Head of Editorial Policy, Nature Research.
