No research article exists in a vacuum. There are no scientific texts in modern science that do not reference other works, and generally those references are meant to support the authors’ assertions. References are a well-understood way to evaluate a piece of research, and those references help tell a story about how the authors reached their conclusions. References tell the story of the research itself; they show the history, they reveal facts that support the assertions, and they lead the reader to the underlying data.
But what about the story behind the evaluation process that brings that research to the public? There is an important story that needs to be told about this part of the life of the research article. The story behind the editorial process—the subplot, so to speak—can help readers and other researchers understand and evaluate the rigor of the peer review and the quality of the publishing process. It would be really useful for readers, funders, and institutions to know what kind of quality checks were made on the manuscript, how many reviewers assessed which versions, and what changes the editor asked the author to make. All of these rich details that are hidden within the editorial workflow should be part of the story.
There is an interesting initiative underway to help tell the story behind the editorial process for any document that claims to be a scholarly work. The initiative, called the DocMap Framework, seeks to define a machine-readable protocol that anyone can use to either communicate the details of the editorial process or to receive and interpret those details. DocMaps has been described as a sort of breadcrumb trail that shows the path that a piece of research, such as a research article, has followed during the evaluation process. Unlike actual breadcrumb trails, DocMaps do not necessarily tell a linear story, since evaluation of an article can have multiple paths that might occur in parallel or in nonsequential ways. It should be noted that this initiative is not intended to be limited to scholarship, and the DocMaps Framework could be used to tell the story of any document that claims to have undergone any sort of vetting, such as a news article or government report.
An editorial story might sound like this: AUTHOR submitted an ARTICLE TYPE called TITLE to JOURNAL on DATE. The article went through SIMILARITY CHECK and REFERENCE CHECK. The EDITOR assigned REVIEWER 1 and REVIEWER 2. REVIEWER 1 said BLAH BLAH BLAH. Reviewer 2 said YADA YADA YADA. EDITOR felt that the paper needed a statistical review, and so she asked STAT-REVIEWER to weigh in. STAT-REVIEWER said that the paper needed to conform to SPECIFIC STATISTICAL METHODOLOGY. EDITOR asked AUTHOR to REVISE.
As humorist John Hodgman says, “Specificity is the soul of narrative,” and in the above story, the words in uppercase would be filled in with the actual details of what evaluation took place during the editorial process.
To be clear, the DocMaps Framework does not tell a story in the narrative way demonstrated above; rather, the DocMaps Framework provides a machine-readable structure in which the details are meaningfully listed (Figure). The DocMap would be embedded in the electronic document, not to be read by the reader, but rather to be received and read by other systems. Those other systems can then take that information and assemble the narrative in whatever format the receiver intends. For example, a journal platform or indexing service might develop a badging system to indicate that certain quality checks were performed. A funding body might use the information to analyze the rigor of peer review, and to draw comparisons between specific types of peer review and article impact. A publisher might use the information to assess their editorial processes in general and make adjustments to increase efficiency.
DocMaps Origin
DocMaps was started by a small group of individuals from the Knowledge Futures Group, a nonprofit which started as a collaboration between MIT Press and MIT Media Lab, ASAPbio (a scientist-driven nonprofit advocating for open communication in the life sciences), and Graz University of Technology (an Austrian public university offering degrees across all technology and natural science disciplines). The DocMaps Framework initiative receives financial support from Howard Hughes Medical Institute. In a press release5 from August of 2020, Gabriel Stein, Jessica Polka, and Tony Ross-Hellauer announced the project as a “new community-endorsed framework for representing editorial research events at the research output level.” In other words, they are seeking to define a mutually agreed method for communicating what milestones a research article has passed through once it is released into the scholarly communications ecosystem.
They noted that they were responding to “numerous efforts to better capture the review processes used on individual articles.” Some of these efforts included the following:
- Transpose (TRANsparency in Scholarly Publishing for Open Scholarship Evolution): a database of peer review journal policies focused on open peer review, coreviewing, and preprints. The goal is to catalog existing policies, to foster experimentation, and to help journals share ideas around peer review.6
- Peer Review Transparency: an initiative to create agreed definitions of how peer review is conducted, and to disclose to readers the kind of review a published scholarly work has gone through. This initiative is supported by the Open Society Foundations.7–9
- Review Maps: an initiative that advocates for the creation of machine-readable “maps” that can be published alongside research articles and other scholarly output. These “maps” would facilitate evaluation of the editorial process by surfacing indicators such as thoroughness and trustworthiness.10
- Peer Review Taxonomy: from the International Association of Scientific, Technical and Medical Publishers (STM), this initiative seeks to standardize definitions and terminology in peer review. STM has stated that, “An agreed peer review taxonomy will help make the peer review process for articles and journals more transparent and will enable the community to better assess and compare peer review practices between different journals.”11
It is also useful to mention 2 other initiatives that seek to create structure around peer review for the purposes of communicating details of the editorial process.
- JATS for Reuse (JATS4R): a working group devoted to optimizing the reusability of scholarly content by developing best practice recommendations for tagging content in JATS XML. The JATS4R Steering Committee reviews recommendations on the use of JATS XML tags that are being proposed by groups working with the National Information Standards Organization (NISO) on related Recommended Practices.12,13
- Manuscript Exchange Common Approach (MECA): a methodology to package files and metadata, including peer review data, in order to transfer that package from one system to any other system, such as from one submission system to another, or from a preprint server to a submission system (and vise versa), or from an authoring system to a preprint server or submission system. Secondarily, MECA can be used to transfer a package of files and data from a submission system to a production vendor. The goal is to have a common process and method to exchange manuscripts in any direction without having unique requirements from system to system. MECA is a NISO Recommended Practice.14
What Is DocMaps?
Each of these initiatives approach the topic from different, usually more narrowly focused perspectives. However, DocMaps is intended to accommodate a wider set of constituents as well as a wider set of use cases. The DocMaps project goes beyond traditional scholarly publishing practices and seeks to also support new, emerging editorial models, like preprints, postpublication review, and open science.
Three requirements were identified as essential for developing a framework that could be used by a constituency as varied as scholarly publishing.
- Extensibility—a wide range of editorial process events should be able to be represented, ranging from a simple assertion that a review occurred to a complete history of editorial comments on a document to a standalone review submitted by an independent reviewer.
- Machine-readability—assertions should be represented in a format that can be interpreted computationally and translated into visual representations.
- Interoperability—a single service should be able to interpret multiple taxonomies against the same criteria and arrive at the same interpretations.
The DocMaps team, which was expanded to also include Gary McDowell, of Lightoller LLC, has produced a white paper,15 which has been posted on the bioRxiv preprint server. This preprint describes the efforts of the DocMaps Technical Committee, of which I was a member. The technical committee met several times over the course of a few months, and using the Delphi Method, this group of 18 people from across scholarly publishing defined 2 use cases on which to focus their efforts.
When examining editorial processes, it becomes clear that there are many variations in workflow. There are various participants (e.g., types of editors and reviewers), QA procedures (e.g., plagiarism and reference checks), and different ways that events are sequenced. In the wake of COVID-19, new paradigms have arisen, such as the increased use of preprints, overlay journals, and presubmission peer review, which meant that the technical committee had to consider what seemed like an ever-expanding array of options. Limiting this initial exploration to just 2 use cases had the practical effect of keeping the conversations focused.
The 2 use cases were as follows:
- A publisher captures context about a review of an article published in their journal.
- An independent review service notifies a preprint server about a review of an article on their platform.
Once the use cases were identified, the technical committee went about creating the actual DocMaps Framework by identifying what events would likely take place, what aspects of those events needed to be described, and how they would be described. Content type schemas were drafted and the resulting proposed DocMaps Framework was posted online16 on January 11, 2021.
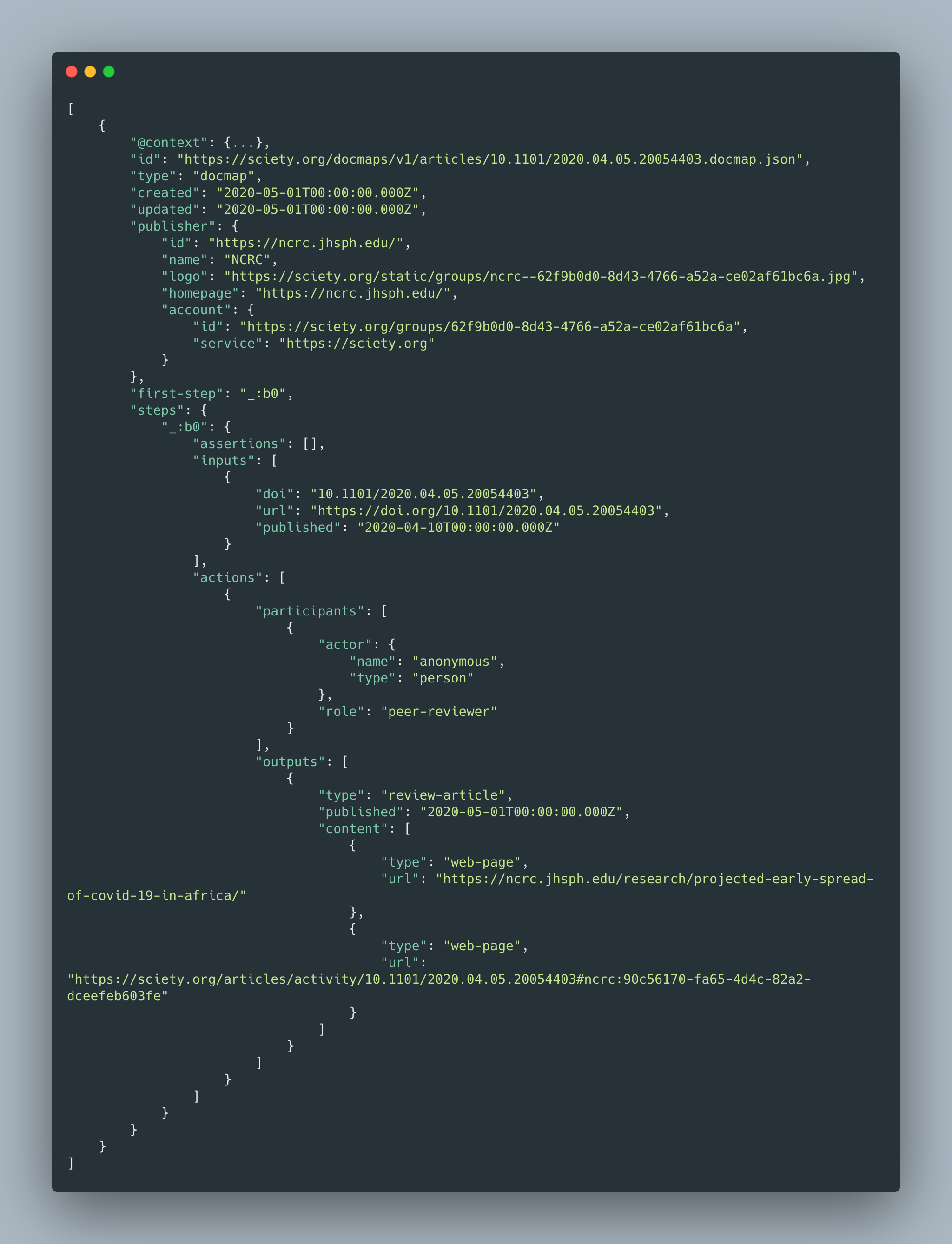
The draft DocMaps Framework document includes sample JavaScript object notation (JSON) along with usage guidance for constructing a DocMap. These are really just examples that have not yet been finalized. The JSON samples for the 2 use cases, as shown in the preprint on bioRxiv, can be found in the Table.
Table. JSON examples of DocMaps.
| 1. “In this example, a journal is describing a double-masked peer review of an article with two rounds of revisions. They do this by nesting a Review context within an Article Context. They then further nest two Version Contexts within the Review Context to describe multiple rounds of feedback.” | { contentType: “article” content: https://doi.org/article/123 createdOn: 2020-08-16T00:00:00Z provider: https://myjournal.org title: ‘An article about something!’ contributors: [ { name: “Liz Jones” id: https://orcid.org/0002-0002 role: “author” } { name: “Eric Mays” id: https://orcid.org/0005-0001 role: “data visualization” } ] datePublished: 2020-01-01T:00:00Z versions: [ { contentType: “version” content: https://doi.org/article/123v1 date_submitted: 2019-12-20T00:00:00Z date_online: 2020-08-15T00:00:00Z ethics_statements: “This was conducted ethically.” ocmpeting_interests: “There were no conflicts of interest.” } ] reviews: [ { contentType: “review” content: https://doi.org/review/abcd createdOn: 2020-06-01T00:00:00z provider: https://myjournal.org decision_date: 2020-07-20T00:00:00z decision: ‘accept with revisions’ contributors: [ { name: “John Doe” affiliation: “Wassamatta U” roles: [editor, author] } { id: 12345 roles: [reviewer] } { id: 23456 roles: [reviewer] } ] identity_transparency: ‘double-anonymized’ reviewer_interacts_with: [editor] review_information_published: [editor-identities] versions: [ { contentType: “version” createdOn: 2020-06-15T00:00:00Z contributors: [ { id: 12345 roles: [reviewer] } { id: 23456 roles: [reviewer] } ] } { contentType: “version” createdOn: 2020-07-10T00:00:00Z date_online: 2020-08-15T00:00:00Z contributors: [ { id: 12345 roles: [reviewer] } ] } ] } ] } |
| 2. “In this example, a review service is describing a fully transparent review of a preprint article with links to the review report and author response. They do this by including a content field for the review object and filling out the author response and STM Association Taxonomy metadata to describe the process of the review.” | { contentType: “review” content: https://doi.org/review/123 createdOn: 2020-08-01T00:00:00z provider: https://myreviewservice.org decision_date: 2020-07-20T00:00:00z decision: “accept” contributors: [ { name: “Tricia McMillan” affiliation: “Maximegalon University” roles: [editor, author] id: https://orcid.org/0000-0000 author_suggested: false }, { name: “Zaphod Beeblebrox” affiliation: “Betelgeuse State College” roles: [reviewer] id: https://orcid.org/0001-0001 author_suggested: true } { name: “Arthur Dent” affiliation: “BBC” roles: [reviewer] id: https://orcid.org/0002-0002 } { name: “Ford Prefect” affiliation: “Pan Galactic Gargle Blaster Society” roles: [invited_reviewer] id: https://orcid.org/0002-0002 } ] author_responses: [{ contentType: “version” content: https://doi.org/response/123 date_online: 2020-07-31T00:00:00Z date_submitted: 2020-07-15T00:00:00Z }] identity_transparency: [all-identities-visible, opt-in] reviewer_interacts_with: [editors, reviewers, authors] review_information_published: [reviewer-identities, editor-identities, review-reports-author-opt-in] versions: [ { contentType: “version” date_submitted: 2020-06-15T00:00:00Z date_online: 2020-07-31T00:00:00Z } ] isReviewOf: [ { contentType: “article” content: https://doi.org/preprint/123 } ] |
Next Steps
There is now an informal working group with members from Knowledge Futures, Cold Spring Harbor Laboratory, eLife’s Sciety, and EMBO’s Early Evidence Base working on a DocMaps pilot implementation focused on use case no. 2. This working group will pilot the DocMaps Framework by applying it to the evaluation of preprints posted on the bioRxiv and medRxiv preprint servers, using evaluations aggregated by Early Evidence Base and Sciety. The intention is to show how DocMaps will provide machine-readable data and context about how community groups and peer review platforms are evaluating preprints.17
Exposing the details and telling the full story behind what goes into preparing a piece of research will help people identify bad, or at least insufficiently evaluated, science. It will also increase trust by confirming that a piece of research has been sufficiently and rigorously vetted. Through embedded code that can be read by any system set up to do so, DocMaps will reveal the inner workings of the editorial process—from technical checks, to peer review, to the editor’s communication with the author. These important details are part of the story that needs to be told. By providing a standardized, machine-readable way to tell the story, downstream systems can take those details and build reports, compare processes, and supplement the research narrative with a subplot about the evaluation process. This means that funders, researchers, journalists, policy makers and readers will have a means by which they can evaluate the rigor of the editorial process. The eventual adoption of the DocMaps Framework means greater transparency in scientific communications and scholarly publishing.
Disclosure
Tony Alves participated on the DocMaps Framework Technical Committee and is co-chair of the NISO MECA Standing Committee.
References and Links
- https://sciety.org/docmaps/v1/articles/10.1101/2020.04.05.20054403.docmap.json
- https://sciety.org/articles/activity/10.1101/2020.04.05.20054403#ncrc:90c56170-fa65-4d4c-82a2-dceefeb603fe
- https://ncrc.jhsph.edu/research/projected-early-spread-of-covid-19-in-africa/
- Pearson CAB, Van Schalkwyk C, Foss AM, O’Reilly KM, SACEMA Modelling and Analysis Response Team, CMMID COVID-19 Working Group, Pulliam JRC. Based on current trends, almost all African countries are likely to report over 1000 COVID-19 cases by the end of April 2020, and over 10,000 a few weeks after that. medRxiv. https://doi.org/10.1101/2020.04.05.20054403
- https://docmaps.knowledgefutures.org/pub/pm6k3fjo/release/1
- https://transpose-publishing.github.io/#/about
- https://www.prtstandards.org/
- https://www.prtstandards.org/pub/zi2i5dt4/release/1
- https://www.opensocietyfoundations.org/
- https://notes.knowledgefutures.org/pub/lgsurhy2/release/1
- https://www.stm-assoc.org/standards-technology/2020-stm-research-data-year/peer-review-taxonomy-project/
- https://jats4r.org/
- https://groups.niso.org/publications/rp/
- https://www.manuscriptexchange.org/
- McDowell GS, Polka JK, Ross-Hellauer T, Stein G. The DocMaps Framework for representing assertions on research products in an extensible, machine-readable, and discoverable format. bioRxiv. https://doi.org/10.1101/2021.07.13.452204
- https://docmaps.knowledgefutures.org/pub/sgkf1pqa/release/4
- https://docmaps.knowledgefutures.org/pub/bwem5bja/release/2
Tony Alves, SVP, Product Management, HighWire Press.
Opinions expressed are those of the authors and do not necessarily reflect the opinions or policies of their employers, the Council of Science Editors or the Editorial Board of Science Editor.
